Publications

119. Phosphate-binding protein-loaded iron oxide particles: Adsorption performance for phosphorus removal and recovery from water
Faten B. Hussein, Andrew H. Cannon, Justin M. Hutchison, Christopher B. Gorman, Yaroslava G. Yingling, Brooke K. Mayer, Environmental Science: Water Research & Technology (2024), DOI:10.1039/D4EW00052H
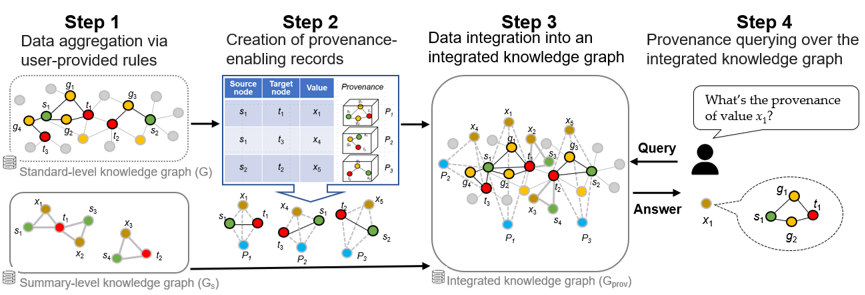
118. BUILD-KG: Integrating Heterogeneous Data Into Analytics-Enabling Knowledge Graphs
Kara Schatz, Pei-Yu Hou, Alexey V. Gulyuk, Yaroslava G. Yingling, and Rada Chirkova, 2023 IEEE International Conference on Big Data (BigData), Sorrento, Italy, 2023 pp. 2965-2974, DOI: 10.1109/BigData59044.2023.10386570
117. Electrostatic self-assembly yields a structurally stabilized PEDOT:PSS with efficient mixed transport and high-performance OECTs
Laine Taussig, Masoud Ghasemi, Sanggil Han, Albert L. Kwansa, Ruipeng Li, Nathan Woodward, Yaroslava G. Yingling, George G. Malliaras, Enrique D. Gomez, Aram Amassian, Matter (2024), DOI:10.1016/j.matt.2023.12.021
- News Release
116. Structural determination of a full-length plant cellulose synthase informed by experimental and in silico methods
Albert L. Kwansa, Abhishek Singh, Justin T. Williams, Candace H. Haigler, Alison W. Roberts, Yaroslava G. Yingling, Cellulose 31 (2024) 1429-1447, DOI:10.1007/s10570-023-05691-x

Selected for Front Cover
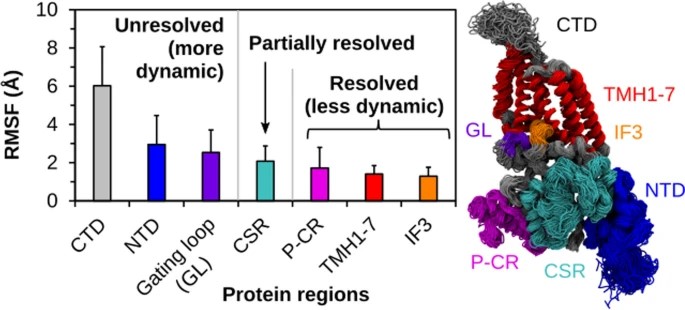
115. Provenance-Aware Data Integration and Summarization Querying for Knowledge Graphs
Pei-Yu Hou, Jing Ao, Kara Schatz, Alexey V Gulyuk, Yaroslava G Yingling, Rada Chirkova, In: Delir Haghighi, P., et al. Information Integration and Web Intelligence. iiWAS 2023. Lecture Notes in Computer Science, vol 14416. Springer, Cham. (2023) DOI:0.1007/978-3-031-48316-5_29
- Best Paper Award at the IIWAS: Information Integration and Web Intelligence Conference 2023, Bali, Indonesia
114. Resolving Structure of ssDNA in Solution by Fusing Molecular Simulations and Scattering Experiments with Machine Learning
Juseok Choi, Albert L. Kwansa, Yaroslava G. Yingling, and Seong H. Kim, J. Phys. Chem B. 127 (2023) 8456-8467 DOI:10.1021/acs.jpcb.3c03910
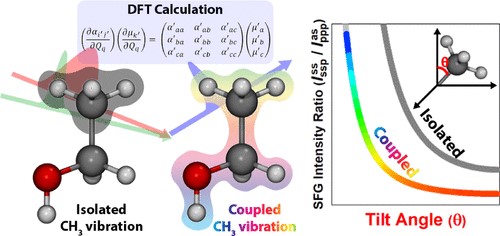
113. Resolving Structure of ssDNA in Solution by Fusing Molecular Simulations and Scattering Experiments with Machine Learning
Thomas J. Oweida and Yaroslava G. Yingling, Advanced Theory and Simulations 6 (2023) 2300411 DOI:10.1002/adts.202300411

Selected for Journal Cover

112. Computer-Assisted Design and Characterization of RNA Nanostructures
 |
Christina J. Bayard and Yaroslava G. Yingling Methods in Molecular Biology (Clifton, NJ) 2709 (2023) 31-49 DOI: 10.1007/978-1-0716-3417-2_2 |
111. Insights into substrate coordination and glycosyl transfer of poplar cellulose synthase-8
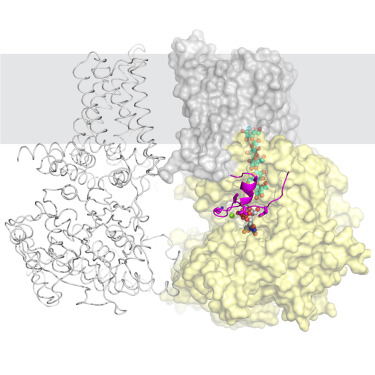 |
Preeti Verma, Ruoya Ho, Albert L. Kwansa, Yaroslava G. Yingling, and Jochen Zimmer Structure 31 (2023) 1-8 DOI: 10.1016/j.str.2023.07.010 |
110. Molecular Mechanism of Plasticizer Exudation from Polyvinyl Chloride
 |
Albert L. Kwansa, Rakhee C. Pani, Joseph A. DeLoach, Arianna Tieppo, Eric J. Moskala, Steven T. Perri, and Yaroslava G. Yingling Macromolecules 56 (2023) 4775-4786 DOI:10.1021/acs.macromol.2c01735 |
109. Solvent Effects in Ligand Stripping Behavior of Colloidal Nanoparticles
 |
Akhlak U. Mahmood, Mehedi H. Rizvi, Joseph B. Tracy, Yaroslava G. Yingling ACS Nano (2023) DOI:10.1021/acsnano.3c01313
|
108. Correlation of Emulsion Chemistry, Film Morphology, and Device Performance in Polyfluorene LEDs Deposited by RIR-MAPLE
 |
Buang Zhang, Sabila K. Pinky, Albert L. Kwansa, Spencer Ferguson, Yaroslava G. Yingling, Adrienne D. Stiff-Roberts ACS Applied Materials & Interfaces 15 (2023) 18153-18165 DOI:10.1021/acsami.3c03012 |
107. Effect of Solvent on the Emulsion and Morphology of Polyfluorene Films: All-atom molecular dynamics approach
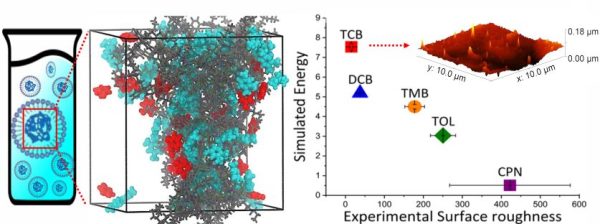 |
Sabila K. Pinky, Albert L. Kwansa, Buang Zhang, Adrienne D. Stiff-Roberts, Yaroslava G. Yingling Soft Matter 19 (2023) 1782-1790 DOI: 10.1039/D2SM01001A |
106. Role of Nanoscale Morphology on the Efficiency of Solvent-Based Desalination Method
 |
James S. Peerless, Alexey V. Gulyuk, Nina J. B. Milliken, Gyu Dong Kim, Elliot Reid, Jae Woo Lee, Dooil Kim, Zachary Hendren, Young Chul Choi, Yaroslava G. Yingling ACS EST Water 3 (2023) 400-409 DOI:10.1021/acsestwater.2c00473 |
105. Squid Skin Cell-Inspired Refractive Index Mapping of Cells, Vesicles, and Nanostructures
 |
Atrouli Chatterjeea, Preeta Pratakshyab, Albert L. Kwansa, Nikhil Kaimal, Andrew Cannon, Barbara Sartori, Benedetta Marmiroli, Helen Orins, Samantha Drake, Justin Couvrette, LeAnn Le, Sigrid Bernstorff, Yaroslava G. Yingling, Alon A. Gorodetsky ACS Biomaterials Science and Engineering 9 (2023) 978-990 DOI:10.1021/acsbiomaterials.2c00088
|
104. Gold Nanoparticle Design for dsRNA Compaction
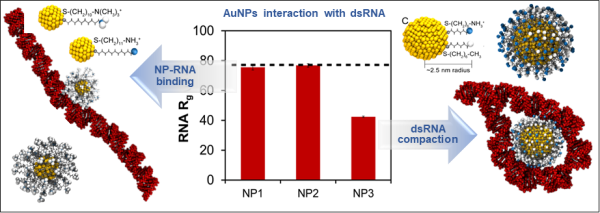 |
Jessica A. Nash, Matthew D. Manning, Alexey V. Gulyuk, Aleksey E. Kuznetsov, Yaroslava G. Yingling Biointerphases 17 (2022) 061001 DOI:10.1116/6.0002043
|
103. Evidence for Plant-Conserved Region Mediated Trimeric CESAs in Plant Cellulose Synthase Complexes
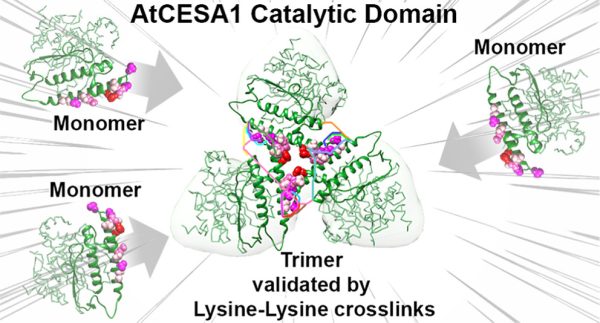 |
Juan Du, Venu Gopal Vandavasi, Kelly R. Molloy, Hui Yang, Lynnicia N. Massenburg, Abhishek Singh, Albert L. Kwansa, Yaroslava G. Yingling, Hugh O’Neill, Brian T. Chait, Manish Kumar, B. Tracy Nixon Biomacromolecules 23 (2022) 3663-3677DOI: 10.1021/acs.biomac.2c00550 |
102. A Comparison between the Lower Critical Solution Temperature Behavior of Polymers and Biomacromolecules
 |
Yuxin Xie, Nan K. Li, Abhishek Singh, Sanket A. Deshmukh, and Yaroslava G. Yingling PhysChem 2 (2022) 52-71 DOI:10.3390/physchem2010005
|
101. All-Atom Simulation Method for Zeeman Alignment and Dipolar Assembly of Magnetic Nanoparticles
 |
Akhlak U. Mahmood and Yaroslava G. Yingling ACS Journal of Chemical Theory and Computations (2022) DOI: 10.1021/acs.jctc.1c01253 |
100. Mapping the Morphological Landscape of Oligomeric Di-block Peptide-Polymer Amphiphiles
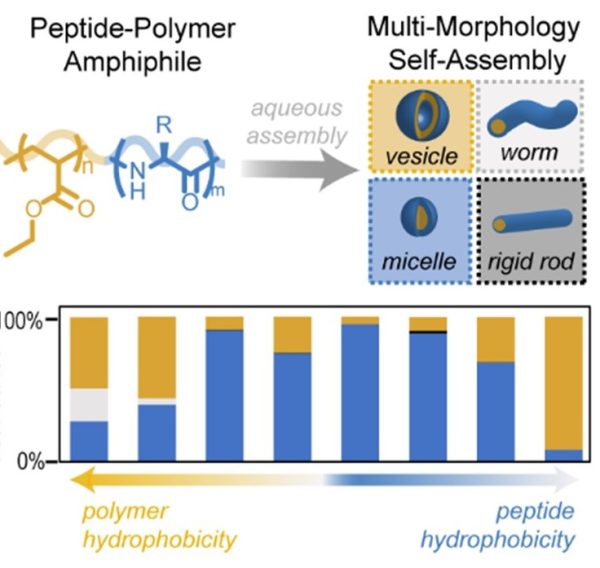 |
Benjamin P. Allen,Zoe M. Wright,Hailey F. Taylor,Thomas J. Oweida,Sabila Kader-Pinky,Emily F. Patteson,Kara M. Bucci,Caleb A. Cox,Abishec Sundar Senthilvel,Yaroslava G. Yingling,Abigail S. Knight Angewandte Chemie (2022) DOI: 10.1002/anie.202115547 |
99. Controlled Organization of Inorganic Materials Using Biological Molecules for Activating Therapeutic Functionalities
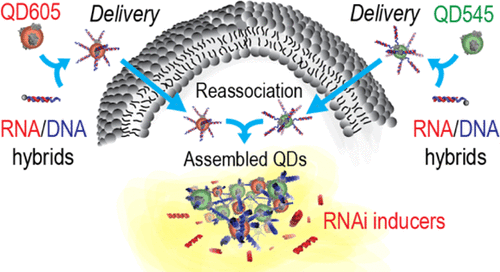 |
Morgan Chandler, Brian Minevich, Brandon Roark, Mathias Viard, M. Brittany Johnson, Mehedi H. Rizvi, Thomas A. Deaton, Seraphim Kozlov, Martin Panigaj, Joseph B. Tracy, Yaroslava G. Yingling, Oleg Gang, Kirill A. Afonin ACS Applied Materials & Interfaces 13 (2021) 39030-39041 DOI: 10.1021/acsami.1c09230 |
98. Insights into Elastin-like Polypeptides Coacervates Structure and Aggregation Behavior: All-atom Molecular Dynamics Simulations
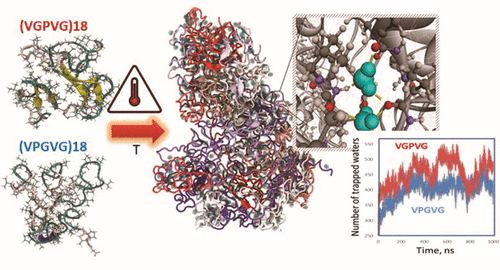 |
Nan K. Li, Yuxin Xie and Yaroslava G. Yingling Journal of Physical Chemistry B 125 (2021) 8627-8635 DOI:10.1021/acs.jpcb.1c02822 |
97. Phenotypic effects of changes in the FTVTxK region of an Arabidopsis secondary wall cellulose synthase compared with results from analogous mutations in other isoforms
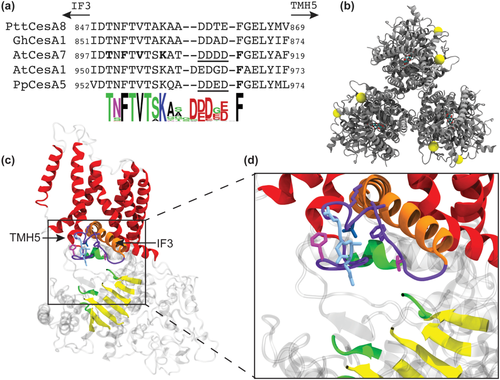 |
Jason N. Burris, Mohamadamin Makarem, Erin Slabaugh, Arielle Chaves, Ethan T. Pierce, Jongcheol Lee, Sarah N. Kiemle, Albert L. Kwansa, Abhishek Singh, Yaroslava G. Yingling, Alison W. Roberts, Seong H. Kim, and Candace H. Haigler Plant Direct 5 (2021) e335 DOI: 10.1002/pld3.335 |
96. Uncertainty Quantification of Atomistic Partial Charges for the Prediction of Solvent Properties via Molecular Dynamics Simulations
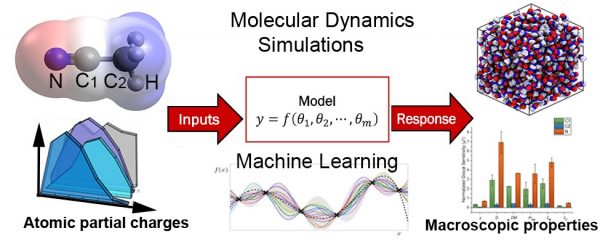 |
James S. Peerless, Albert L. Kwansa, Branden S. Hawkins, Ralph C. Smith, Yaroslava G. Yingling ACS Journal of Chemical Information and Modeling 61 (2021) 1745-1761 DOI: 10.1021/acs.jcim.0c01204 |
95. Weakly Ionically Bound Thermo-sensitive Hyperbranched Polymers
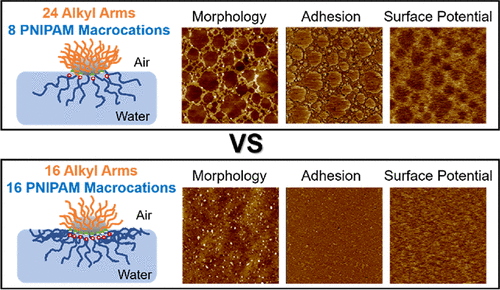 |
Hansol Lee, Aleksandr Stryutsky, Akhlak-Ul Mahmood, Abhishek Singh, Valery V. Shevchenko, Yaroslava G. Yingling, Vladimir V. Tsukruk Langmuir 37 (2021) 2913-2927 DOI: 10.1021/acs.langmuir.0c03487 |
94. Assessment of AMBER Force Fields for Simulations of ssDNA Structure
 |
Thomas J. Oweida, Ho Shin Kim, Abhishek Singh, and Yaroslava G. Yingling ACS Journal of Chemical Theory and Computation 17 (2021) 1208-1217 DOI: 10.1021/acs.jctc.0c00931 |
93. Structure, self-assembly, and properties of a truncated reflectin variant
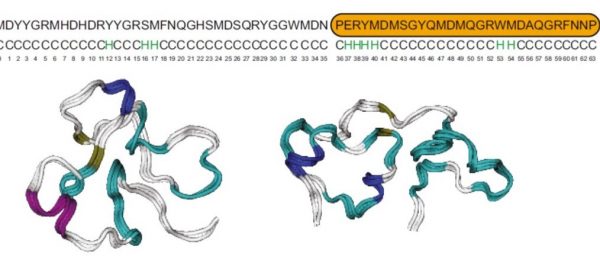 |
Mehran J. Umerani, Preeta Pratakshya, Atrouli Chatterjee, Juana A. Cerna Sanchez, Ho Shin Kim, Gregor Ilc, Matic Kovačič, Christophe Magnan, Benedetta Marmiroli, Barbara Sartori, Albert L. Kwansa, Helen Orins, Andrew W. Bartlett, Erica M. Leung, Zhijing Feng, Kyle L. Naughton, Brenna Norton-Baker, Long Phan, James Long, Alex Allevato, Jessica E. Leal-Cruz, Qiyin Lin, Pierre Baldi, Sigrid Bernstorff, Janez Plavec, Yaroslava G. Yingling, and Alon A. Gorodetsky Proceedings of the National Academy of Sciences of the United States of America 117 (2020) 32891-32901 DOI: 10.1073/pnas.2009044117 |
92. Co-assembling Polysaccharide Nanocrystals and Nanofibers for Robust Chiral Iridescent Films
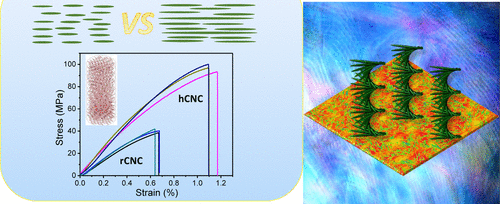 |
Rui Xiong, Abhishek Singh, Shengtao Yu, Shuaidi Zhang, Hansol Lee, Yaroslava G. Yingling, Dhriti Nepal, Timothy J. Bunning, Vladimir V. Tsukruk ACS Applied Materials & Interfaces 12 (2020) 35345-35353 DOI: 10.1021/acsami.0c08571 |
91. Partially fluorinated copolymers as oxygen sensitive 19F MRI agents
 |
Nicholas G Taylor; Sang Hun Chung; Albert L Kwansa; Robert R Johnson III; Aaron J Teator; Nina J B Milliken; Karl M Koshlap; Yaroslava G Yingling; Yueh Z Lee; Frank Leibfarth Chemistry – A European Journal 26 (2020) 9982-9990 DOI: 10.1002/chem.202001505 |
90. In silico structure prediction of full-length cotton cellulose synthase protein (GhCESA1) and its hierarchical complexes
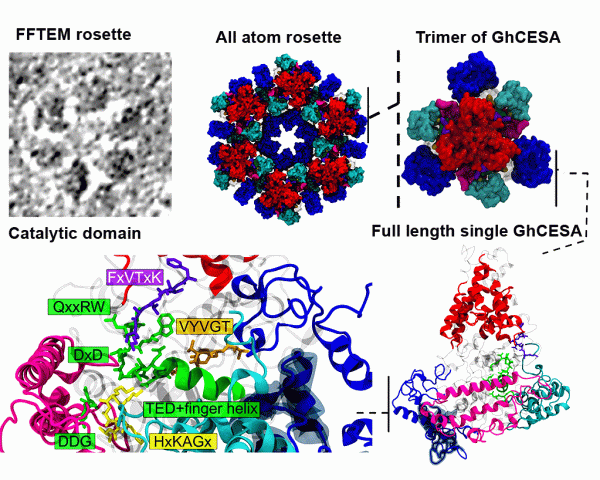 |
Abhishek Singh, Albert L. Kwansa, Ho Shin Kim, Justin T. Williams, Hui Yang, Nan K. Li, James D. Kubicki, Alison W. Roberts, Candace H. Haigler, Yaroslava G. Yingling Cellulose 27 (2020) 5597-5616 DOI: 10.1007/s10570-020-03194-7 |
89. Anisotropic optical and frictional properties of Langmuir–Blodgett film consisting of uniaxially-aligned rod-shaped cellulose nanocrystals
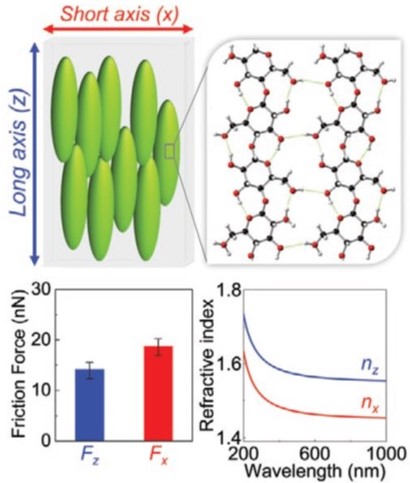 |
Inseok Chae, Dien Ngo, Zhe Chen , Albert L. Kwansa, Xing Chen, Amira Barhoumi Meddeb, Nikolas J. Podraza, Yaroslava G. Yingling, Zoubeida Ounaies and Seong H. Kim Advanced Materials Interfaces 7 (2020) 1-18 DOI:10.1002/admi.201902169 |
88. Merging Materials and Data Science: Opportunities, Challenges, and Education in Materials Informatics
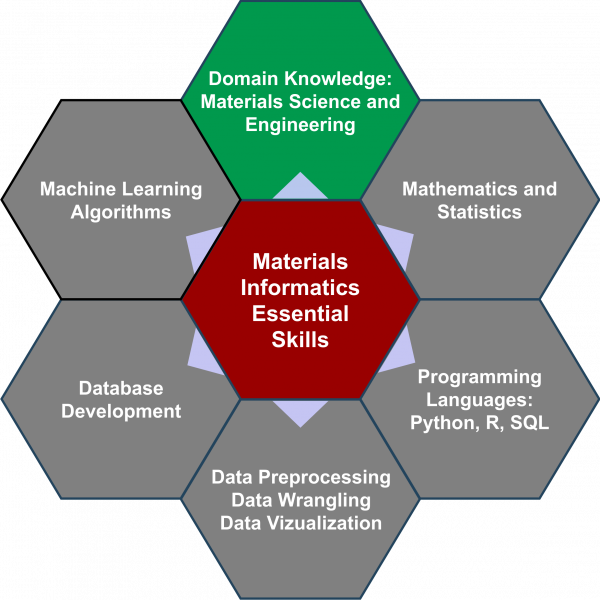 |
Thomas J. Oweida, Akhlak Ul-Mahmood, Matthew D. Manning, Sergei Rigin, Yaroslava G. Yingling MRS Advances 5 (2020) 1-18 DOI: 10.1557/adv.2020.171 |
87. Effect of octadecylamine surfactant concentration on DNA structure on graphene surfaces
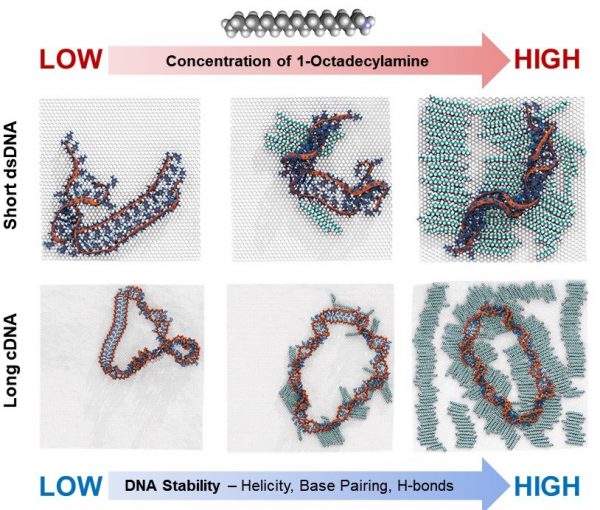 |
Ho Shin Kim, Nathanael A. Brown, Stefan Zauscher, and Yaroslava G. Yingling Langmuir 36 (2020) 931-938 DOI:10.1021/acs.langmuir.9b02926 |
86. Materials matter in phosphorus sustainability
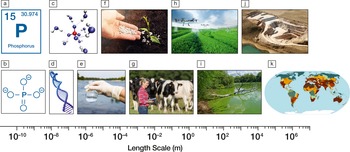 |
Jacob L. Jones, Yaroslava G. Yingling, Ian M. Reaney, and Paul Westerhoff MRS Bulletin 5 (2020) 45(1), 7-10 DOI:10.1557/mrs.2020.4 |
85. Intrinsically disordered proteins access a range of hysteretic phase separation behaviors
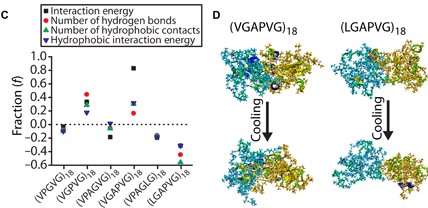 |
Felipe Garcia Quiroz, Nan K. Li, Stefan Roberts, Patrick Weber, Michael Dzuricky, Isaac Weitzhandler, Yaroslava G. Yingling and Ashutosh Chilkoti Science Advances 5 (2019) eaax5177 DOI: 10.1126/sciadv.aax5177 |
84. Biochemical and physiological flexibility accompanies reduced cellulose biosynthesis in Brachypodium cesa1 S830N
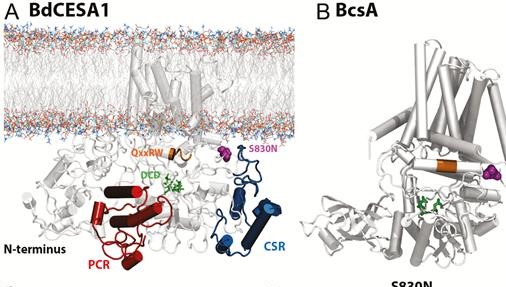 |
Chad Brabham, Abhishek Singh, Jozsef Stork, Ying Rong, Indrajit Kumar, Kazuhiro Kikuchi, Yaroslava G Yingling, Thomas P Brutnell, Jocelyn K C Rose, Seth DeBolt AoB PLANTS 11 (2019) plz041 DOI:10.1093/aobpla/plz041 |
83. Simulations of Cellulose Synthesis Initiation and Termination in Bacteria
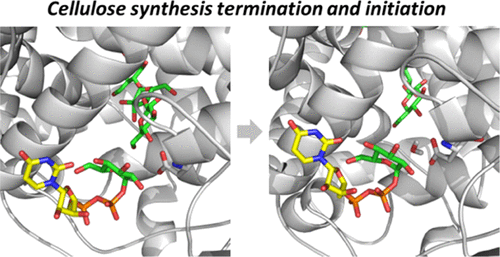 |
Hui Yang, John B. McManus, Daniel Oehme, Abhishek Singh, Yaroslava G Yingling, Ming Tien, and James D. Kubicki J. Phys. Chem. B 123 (2019) 3699-3705 DOI:10.1021/acs.jpcb.9b02433 |
82. Dissipative Particle Dynamics Approaches to Modeling the Self-Assembly and Morphology of Neutral and Ionic Block Copolymers in Solution
 |
Thomas A. Deaton, Fikret Aydin, Nan K. Li, Xiaolei Chu, Meenakshi Dutt and Yaroslava Yingling Molecular Modeling and Simulation, Applications and Perspectives, Series Ed.: E. Maginn (2019) book chapter |
81. High-Performance Chromatographic Characterization of Surface Chemical Heterogeneities of Fluorescent Organic–Inorganic Hybrid Core–Shell Silica Nanoparticles
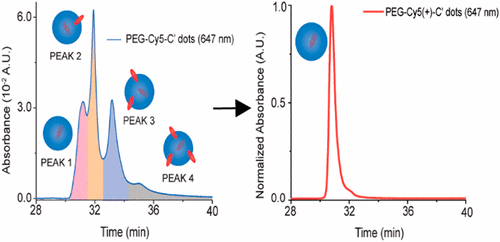 |
Thomas C. Gardinier, Ferdinand F. E. Kohle, James S. Peerless, Kai Ma, Melik Z. Turker, Joshua A. Hinckley, Yaroslava G. Yingling, and Ulrich Wiesner ACS Nano 13 (2019) 1795-1804 DOI: 10.1021/acsnano.8b07876 |
80. Progress in ligand design for monolayer-protected nanoparticles for nano-bio interfaces
 |
Matthew D. Manning, Albert L. Kwansa, Thomas Oweida, James Peerless, Abhishek Singh, Yaroslava G. Yingling Biointerphases 13 (2018) 06D502 DOI: 10.1116/1.5044381
|
79. Soft Matter Informatics: Current Progress and Challenges
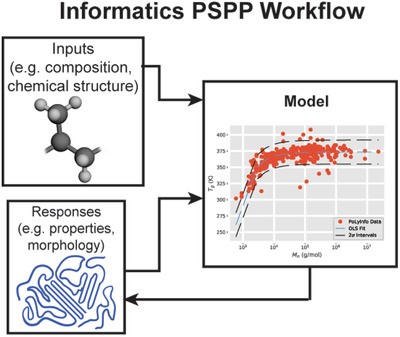 |
James S. Peerless, Nina J. B. Milliken, Matthew D. Manning, Thomas J. Oweida, Yaroslava G. Yingling Advanced Theory and Simulations(2018) DOI: 10.1002/adts.201800129
|
78. Bathophenanthroline Disulfonate Ligand-induced Self-assembly of Ir(III) Complexes in Water: An Intriguing Class of Photoluminescent Soft Materials
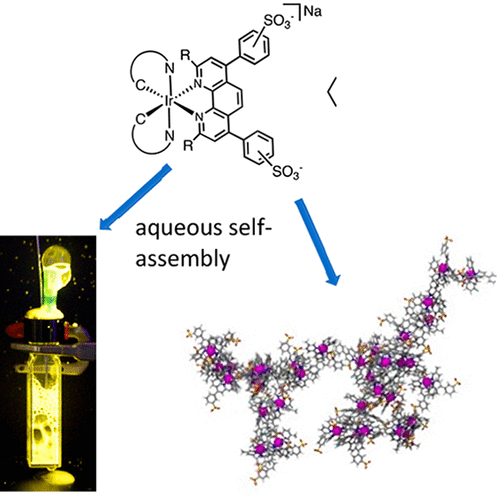 |
Michelle M. McGoorty, Abhishek Singh, Thomas A. Deaton, Benjamin Peterson, Chelsea M. Taliaferro, Yaroslava G. Yingling, and Felix N. Castellano ACS Omega (2018) 14027-14038 DOI: 10.1021/acsomega.8b02034 |
77. Enzymatic Synthesis of Nucleobase-Modified Single-Stranded DNA Offers Tunable Resistance to Nuclease Degradation
 |
Renpeng Gu, Thomas Oweida, Yaroslava G. Yingling, Ashutosh Chilkoti, Stefan Zauscher Biomacromolecules (2018) DOI: 10.1021/acs.biomac.8b00816 |
76. Sequence directionality dramatically affects LCST behavior of elastin-like polypeptides
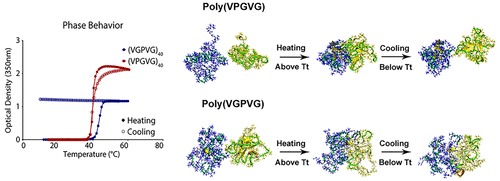 |
Nan K. Li, Stefan Roberts, Felipe Garcia Quiroz, Ashutosh Chilkoti, and Yaroslava G. Yingling Biomacromolecules (2018) DOI: 10.1021/acs.biomac.8b00099 |
75. Wrapping Nanocellulose Nets around Graphene Oxide Sheets
 |
Rui Xiong, Ho Shin Kim, Lijuan Zhang, Volodymyr F. Korolovych, Shuaidi Zhang, Yaroslava G. Yingling, and Vladimir V. Tsukruk Angewandte Chemie International Edition 57 (2018) 8508-8513 DOI:10.1002/anie.201803076 |
74. Cellulose synthase ‘class specific regions’ are intrinsically disordered and functionally undifferentiated
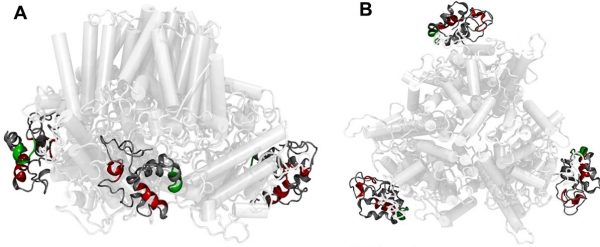 |
Tess R. Scavuzzo-Duggan, Arielle M. Chaves, Abhishek Singh, Latsavongsakda Sethaphong, Erin Slabaugh, Yaroslava G. Yingling, Candace H. Haigler, and Alison W. Roberts Journal of Integrative Plant Biology 53 (2018) 5766-5776 DOI:10.1002/anie.201803076 |
73. Interfacial Stability of Graphene-based Surfaces in water and organic solvents
 |
Ho Shin Kim, Thomas J. Oweida, and Yaroslava G. Yingling Journal of Materials Science (Invited Special Issue) 53 (2018) 5766–5776 DOI: 10.1007/s10853-017-1893-9
|
72. Functional Modification of Silica through Enhanced Adsorption of Elastin-Like Polypeptide Block Copolymers
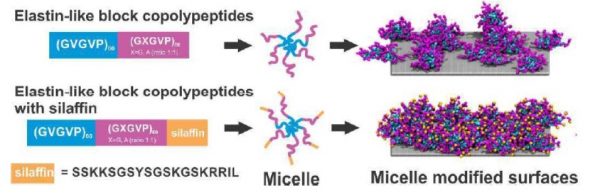 |
Linying Li, Nan K Li, Qing Tu, Owen Im, Chia-Kuei Mo, Wei Han, William H. Fuss, Nick J. Carroll, Ashutosh Chilkoti, Yaroslava G. Yingling, Stefan Zauscher and Gabriel P. López Biomacromolecules (2018) DOI: 10.1021/acs.biomac.7b01307 |
71. Template-Guided Assembly of Silk Fibroin on Cellulose Nanofibers for Robust Nanostructures with Ultrafast Water Transport
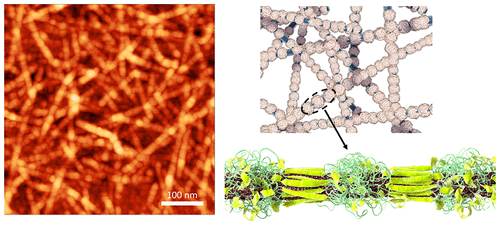 |
Rui Xiong, Ho Shin Kim, Shuaidi Zhang, Sunghan Kim, Volodymyr F. Korolovych, Ruilong Ma, Yaroslava G. Yingling, Canhui Lu, and Vladimir V. Tsukruk ACS Nano (2017) DOI: 10.1021/acsnano.7b04235 |
70. Salt responsive morphologies of ssDNA-based triblock polyelectrolytes in semi-dilute regime: effect of volume fractions and polyelectrolyte length
 |
Nan K. Li, Huihui Kuang, William H. Fuss, Stefan Zauscher, Efrosini Kokkoli and Yaroslava G. Yingling Macromolecular Rapid Communications 38 (2017) 1700422 DOI: 10.1002/marc.201700422 |
69. Effect of C60 Adducts on the Dynamic Structure of Aromatic Solvation Shells
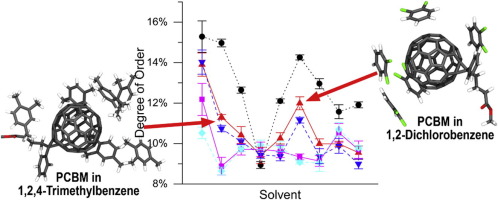 |
James S. Peerless, G. Hunter Bowers, Albert L. Kwansa, Yaroslava G. Yingling Chemical Physics Letters 678 (2017) 79-84 DOI: 10.1016/j.cplett.2017.04.010 |
68. Interfacial Mechanical Properties of Graphene on Self-Assembled Monolayers: Experiments and Simulations
 |
Qing Tu, Ho Shin Kim, Thomas J. Oweida, Zehra Parlak, Yaroslava G Yingling, and Stefan Zauscher ACS Applied Materials and Interfaces 9 (2017) 10203-10213 DOI: 10.1021/acsami.6b16593 |
67. Effect of graphene oxidation rate on adsorption of poly-thymine single stranded DNA
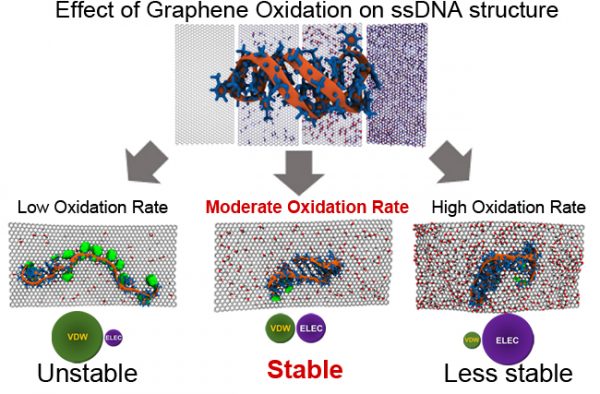 |
Hoshin Kim, Barry L. Farmer, and Yaroslava G. Yingling Advanced Materials Interfaces (2017) DOI: 10.1002/admi.201601168 |
66. Advances in Molecular Modeling of Nanoparticle-Nucleic Acid Interfaces
 |
Jessica A. Nash, Albert L. Kwansa, James S. Peerless, Hoshin Kim, Yaroslava G. Yingling Bioconjugate Chemistry 28 (2017) 3-10 DOI: 10.1021/acs.bioconjchem.6b00534 |
65. Fluorescence blinking as an output signal for biosensing
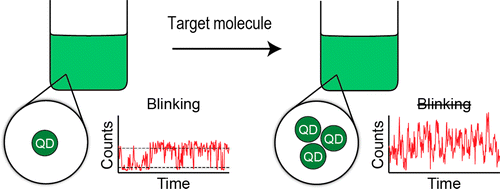 |
Brandon Roark, Jenna A. Tan, Anna Ivanina, Morgan Chandler, Jose Castaneda, Ho Shin Kim, Shriram Jawahar, Mathias Viard, Strahinja Talic, Kristin L. Wustholz, Yaroslava G Yingling, Marcus Jones, and Kirill A Afonin ACS Sensors 1 (2016) 1295-1300 DOI: 10.1021/acssensors.6b00352 |
64. Binding of single stranded nucleic acids to cationic ligand functionalized gold nanoparticles
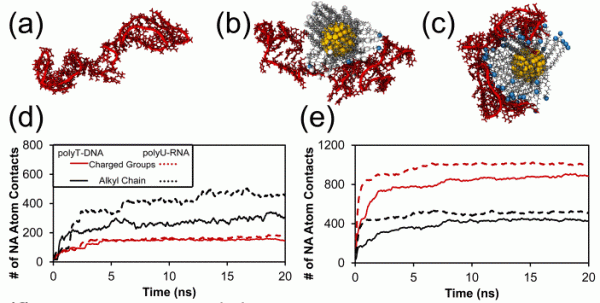 |
Jessica A. Nash, Tasha L. Tucker, William Therriault, Yaroslava G. Yingling Biointerphases 11 (2016) 04B305 DOI: 10.1116/1.4966653 |
63. Silk Fibroin-Substrate Interactions at Heterogeneous Nanocomposite Interfaces
 |
Anise M. Grant, Ho Shin Kim, Trisha L. Dupnock, Kesong Hu, Yaroslava G. Yingling, Vladimir V. Tsukruk Advanced Functional Materials 26 (2016) 6380-6392 DOI: 10.1002/adfm.201601268 |
62. Emulsion-based RIR-MAPLE deposition of conjugated polymers: primary solvent effect and its implications on organic solar cell performance
 |
Wangyao Ge, Nan K. Li, Ryan D. McCormick, Eli Lichtenberg, Yaroslava G. Yingling and Adrienne D. Stiff-Roberts ACS Applied Materials and Interfaces 8 (2016) 19494-19506 DOI: 10.1021/acsami.6b05596 |
61. Comparative structural and computational analysis supports eighteen cellulose synthases in the plant cellulose synthesis complex
 |
B. Tracy Nixon, Katayoun Mansouri, Abhishek Singh, Juan Du, Jonathan K. Davis, Jung-Goo Lee, Erin Slabaugh, Venu Gopal Vandavasi, Hugh O’Neill, Eric. M. Roberts, Alison W. Roberts, Yaroslava G. Yingling, Candace H. Haigler Scientific Reports 6 (2016) 28696 DOI:10.1038/srep28696 |
60. Role of Imidazolium Cation on Structure and Activity of Candida antarctica Lipase B Enzyme in Ionic Liquids
 |
Ho Shin Kim, Doyoung Eom, Yoon-Mo Koo, and Yaroslava G. Yingling Physical Chemistry Chemical Physics 18 (2016) 22062-22069 DOI: 10.1039/C6CP02355J |
59. Sequence dependent interaction of single stranded DNA with graphitic flakes: atomistic molecular dynamics simulations
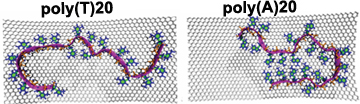 |
Ho Shin Kim, Sabrina M. Huang, Yaroslava G. Yingling MRS Advances 1(2016) 1883-1889 DOI: 10.1557/adv.2016.91 |
58. Design of potent and controllable anticoagulants using DNA aptamers and nanostructures
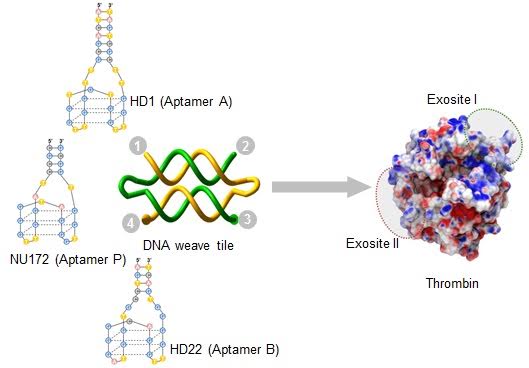 |
Abhijit Rangnekar, Jessica A Nash, Bethany Goodfred, Yaroslava G. Yingling, and Thomas LaBean Molecules 21 (2016) 202 DOI: 10.3390/molecules21020202 |
57. LCST Behavior is Manifested in a Single Molecule: Elastin-like polypeptide (VPGVG)n
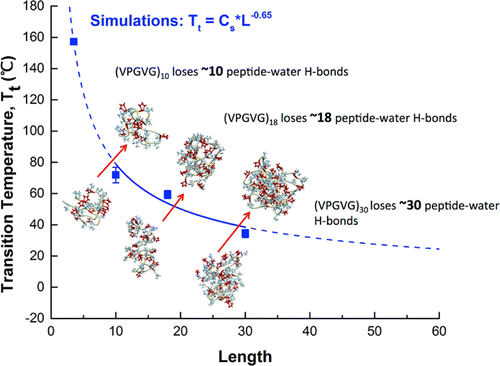 |
Binwu Zhao, Nan K. Li, Yaroslava G. Yingling, and Carol K. Hall Biomacromolecules 17 (2016) 111-118 DOI: 10.1021/acs.biomac.5b01235 |
56. Prediction of the structures of the plant-specific regions of vascular plant cellulose synthases and correlated functional analysis
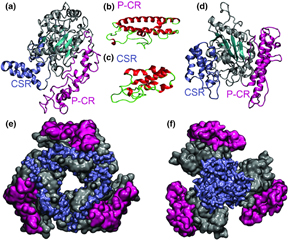 |
Latsavongsakda Sethaphong, Jonathan K. Davis, Erin Slabaugh, Abhishek Singh, Candace H. Haigler, Yaroslava G. Yingling Cellulose 23 (2016) 145-161 DOI: 10.1007/s10570-015-0789-6 |
55. Na+ Dependent Anion Transport by a Barley Efflux Protein Revealed through an Integrative Platform
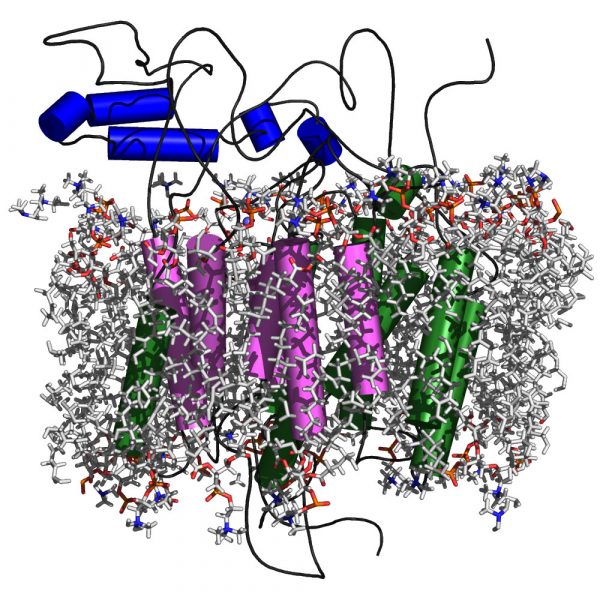 |
Yagnesh Nagarajan, Jay Rongala, Sukanya Luang, Abhishek Singh, Nadim Shadiac, Julie Hayes, Tim Sutton, Matthew Gilliham, Stephen Tyerman, Gordon McPhee, Nicolas H. Voelcker, Hayden D. T. Mertens, Nigel Kirby, Jung-Goo Lee, Yaroslava G. Yingling, Maria Hrmova Plant Cell 28 (2016) 202-218 DOI: 10.1105/tpc.15.00625
|
54. Fullerenes in Aromatic Solvents: Correlation between Solvation Shell Structure, Solvate Formation, and Solubility
 |
James S. Peerless, G. Hunter Bowers, Albert L. Kwansa, and Yaroslava G. Yingling J. Phys. Chem. B 119 (2015) 1534415352 DOI: 10.1021/acs.jpcb.5b09386 |
53. Characterization of nucleic acid compaction with histone-mimic nanoparticles through all-atom molecular dynamics
 |
Jessica A. Nash, Abhishek Singh, Nan K. Li, and Yaroslava G. Yingling ACS Nano 9 (2015) 12374-12382 DOI: 10.1021/acsnano.5b05684
|
52. Microtubules and cellulose biosynthesis: the emergence of new players
 |
Shundai Li, Lei Lei, Yaroslava G. Yingling, and Ying Gu Current Opinion in Plant Biology 28 (2015) 76-82 DOI: 10.1016/j.pbi.2015.09.002 |
51. Cellulose synthase interactive 1 is required for a fast recycling of cellulose synthase complexes to the plasma membrane in Arabidopsis
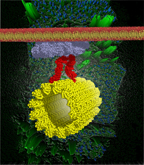 |
Lei Lei, Abhishek Singh, Logan Bashline, Shundai Li, Yaroslava G. Yingling, and Ying Gu Plant Cell (2015) DOI: 10.1105/tpc.15.00442 |
50. Prediction of solvent-induced morphological changes of polyelectrolyte diblock copolymer micelles
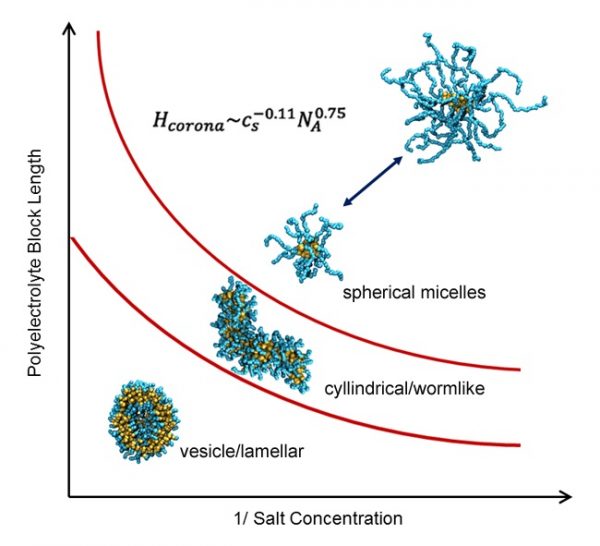 |
Nan K. Li, William H. Fuss, L. Tang, R. Gu, A. Chilkoti, S. Zauscher, Yaroslava G. Yingling Soft Matter 11 (2015) 8236-8245 DOI: 10.1039/C5SM01742D |
49. How Cellulose Elongates – a QM/MM Study of the Molecular Mechanism of Cellulose Polymerization in Bacterial CESA
H. Yang, J. Zimmer, Y.G. Yingling, J. D. Kubicki
Journal of Physical Chemistry B 119 (2015) 6525-6535
48. An Implicit Solvent Ionic Strength (ISIS) Method to Model Polyelectrolyte Systems with Dissipative Particle Dynamics
N. K. Li, W. H. Fuss, Y. G. Yingling
Macromolecular Theory and Simulations 24 (2015) 7-12.
-
-
- Reported in Materials today, Science Daily
- Cover Image
- Video
-
47. Molecular description of the LCST behavior of an elastin-like poly-peptide
N. K. Li, F. García Quiroz, C. K. Hall, A. Chilkoti, Y. G. Yingling
Biomacromolecules 15 (2014) 3522-3530
46. Computational and genetic evidence that different structural conformations of a non-catalytic region affect the function of plant cellulose synthase
E. Slabaugh, L. Sethaphong, C. Xiao, J. Amick, C. T. Anderson, C. H. Haigler, Y. G. Yingling
Journal of Experimental Botany 65 (2014) 6645-6653.
45. Progress in molecular modeling of DNA materials
N. K. Li, H. S. Kim, J. A. Nash, M. Lim, Y. G. Yingling
Molecular Simulation 40 (2014) 777-783.
44. Enzymatic polymerization of high molecular weight DNA amphiphiles that self-assemble into star-like micelles
L. Tang, V. Tjong, N. Li, Y. G. Yingling, A. Chilkoti, S. Zauscher
Advanced Materials 26 (2014) 3050-3054.
43. The relationship between enhanced enzyme activity and structural dynamics in ionic liquids: a combined computational and experimental study
H. S. Kim, S. H. Ha, L. Sethaphong, Y.-M. Koo, Y. G. Yingling
Physical Chemistry Chemical Physics 16 (2014) 2944-2953.
42. Cellulose synthases: new insights from crystallography and modeling
E. Slabaugh, J. K. Davis, C. H. Haigler, Y. G. Yingling, J. Zimmer
Trends in Plant Science 19 (2013) 99-106.
41. Correlating fullerenes diffusion with polythiophene morphology: molecular dynamics simulations
R. C. Pani, B. D. Bond, G. Krishnan, Y. G. Yingling
Soft Matter 9 (2013) 10048-10055
40. Tertiary model of a plant cellulose synthase
L. Sethaphong, C. H. Haigler, J. D. Kubicki, J. Zimmer, D. Bonetta, S. DeBolt, Y. G. Yingling
Proc. Natl. Acad. Sci. USA 110 (2013) 7512-7517
39. X3DBio2: A Visual Analysis Tool for Biomolecular Structure Comparison
H. Yi, S. Thakur, L. Sethaphong, Y. G. Yingling
Proc. SPIE 8654, VDA (2013) 86540W doi:10.1117/12.2002626
-
-
- Download X3DBio2 software
-
38. The effect of point mutations on structure and mechanical properties of collagen-like fibril: a molecular dynamics study
A. E. Marlowe, A. Singh, Y. G. Yingling
Materials Science and Engineering C 32 (2012) 2583-2588
37. Weakly Charged Cationic Nanoparticles Induce DNA Bending and Strand Separation
J. G. Railsback, A. Singh, R. C. Pearce, T. E. McKnight, R. Collazo, Z. Sitar, Y. G. Yingling, A. Melechko
Advanced Materials 24 (2012) 4261-4265
-
-
- Reported in Materials360, nanowiki
- Cover Image
-
36. Role of Solvent and Dendritic Architecture on the Redox Core Encapsulation
R. C. Pani, Y. G. Yingling
J. Phys. Chem. A 116 (2012) 7593-7599
35. X3DBio: A Visual Analysis Tool for Biomolecular Structure Exploration
H. Yi, A. Singh, Y. G. Yingling
Proc. SPIE 8294 (2012) 82940S-1-8
-
-
- Download X3DBio software
-
34. The role of hydrogen bonding in water-mediated glucose solubility in ionic liquids
H. S. Kim, R. Pani, S. H. Ha, Y.-M. Koo, Y. G. Yingling
Journal of Molecular Liquids 166 (2012) 25-30
33. Molecular dynamics simulaitons of bio-nano materials
M. A. Pasquinelli, Y. G. Yingling
Encyclopedia of Nanotechnology (2012) pp.1454-1463
32. Theoretical perspective on properties of DNA-functionalized surfaces
A. Singh, H. Eksiri, Y. G. Yingling
Journal of Polymer Science B: Polymer Physics 49 (2011) 1563-1627
31. Interactions of cations with RNA loop-loop complexes
A. Singh, L. Sethaphong, Y. G. Yingling
Biophysical Journal 101 (2011) 727
30. Simulations of stretching single stranded DNA
A. Singh, Y. G. Yingling
Mater. Res. Soc. Proc. (2011) 1301, mrsf10-1301-oo10-06 doi:10.1557/opl.2011.569.
29. Effect of Oligonucleotide Length on the Assembly of DNA Materials: Molecular Dynamics Simulations of Layer-by-Layer DNA Films
A. Singh, S. Snyder, L. Lee, A. P. R. Johnston, F. Caruso, Y. G. Yingling
Langmuir 26 (2010) 17339-17347
-
-
- Reported in Materials360®, Foresight Institute
-
28. The sequence of HIV-1 TAR RNA helix controls cationic distribution
L. Sethaphong, A. Singh, A. E. Marlowe, Y. G. Yingling
J. Phys. Chem. C 114 (2010) 5506-5512
27. Comparing ion distributions around RNA and DNA helical and loop-loop motifs
A. V. Semichaevsky, A. E. Marlowe, Y. G. Yingling
Mater. Res. Soc. Symp. Proc. Vol. 1130 (2009) W05-05
- A. Singh and Y. G. Yingling, “RNA molecular recognition and self-assembly into nanoparticles”, Proc. International Institute of Informatics and Systemics 1 (2009) 6-7. ISBN: 193427268X;978-193427268-8
- 25. S. Snyder and Y. G. Yingling, “Computational Elucidation of the self-assembly of nucleic acids into novel materials”, Proc. International Institute of Informatics and Systemics 1 (2009) 8-9. ISBN: 193427268X;978-193427268-8
24. The 3′ Proximal Translational Enhancer of Turnip Crinkle Virus Binds to 60S Ribosomal Subunits
V. A. Stupina, A. Meskauskas, J. C. Mccormack, Y. G. Yingling, B. A. Shapiro, J. D. Dinman, A. E. Simon
RNA 14 (2008) 2379-2393
23. Structural Domains Within the 3′ UTR of Turnip Crinkle Virus
J. C. McCormack, X. Yuan, Y. G. Yingling, W. Kasprzak, R. E. Zamora, B. A. Shapiro, A. E. Simon
Journal of Virology 82 (2008) 8706-8720
22. Protocols for the In silico Design of RNA Nanostructures
B. A. Shapiro, E. Bindewald, W. Kasprzak, Y. G. Yingling
Nanostructure Design Methods and Protocols, Methods in Molecular Biology series Vol. 474, E. Gazit and R. Nussinov, Eds., (Humana Press, 2008)
21. RNAJunction: a database of RNA junctions and kissing loops for three-dimensional structural analysis and nanodesign
E.Bindewald, R. Hayes, Y. G. Yingling, W. Kasprzak, B. A. Shapiro
Nucleic Acids Res. 36 (2008) D392-D397
-
-
- Link to RNAJunction database
-
20. Computational design of an RNA hexagonal nano-ring and an RNA nanotube
Y. G. Yingling, B. A. Shapiro
Nano Letters 7 (2007) 2328-2334
19. Bridging the gap in RNA structure prediction
B. A. Shapiro, Y. G. Yingling, W. Kasprzak, E. Bindewald
Current Opinion in Structural Biology 17 (2007) 157-165
18. Incorporation of chemical reactions into UV photochemical ablation of coarse-grained material
Y. G. Yingling, B. J. Garrison
Applied Surface Science 253 (2007) 6377-6381
17. Computational Investigation into the Mechanisms of UV Ablation of Poly (methyl methacrylate)
M. Prasad, P. F. Conforti, B. J. Garrison, Y. G. Yingling
Applied Surface Science 253 (2007) 6382-6385
16. The Impact of Dyskeratosis Congenita Mutations on Structure and Dynamics of the Telomerase RNA Pseudoknot Domain
Y. G. Yingling, B. A. Shapiro
J. Biomol. Str. Dynam. 24 (2007) 303-320
15. Computational studies of ultraviolet ablation of poly(methyl methacrylate)
P. F. Conforti, Y. G. Yingling and B. J. Garrison
Journal of Physics: C 59 (2007) 322-327
14. The Prediction of the Wild-type Telomerase RNA Pseudoknot Structure and the Pivotal Role of a Bulge in its Formation
Y. G. Yingling, B. A. Shapiro
J Mol Graph Model 25 (2006) 261-274
13. Structural and Dynamical Classification of RNA Single-Base Bulges for Nanostructure Design
W. A. Hastings, Y. G. Yingling, G. S. Chirikjian, B. A. Shapiro
J Comp Theor Nanoscience 3 (2006) 63-77
12. Coarse-Grained Model of the Interaction of Light with Polymeric Material: Onset of Ablation
Y. G. Yingling, B. J. Garrison
Journal of Physical Chemistry B 109 (2005) 16482-16489
11. Dynamic Behavior of the Telomerase RNA Hairpin Structure and Its Relationship to Dyskeratosis Congenita
Y. G. Yingling, B. A. Shapiro
Journal of Molecular Biology 348 (2005) 27
10. Theoretical Investigation of Laser Pulse Width Dependence in Thermal Confinement Regime
Y. G. Yingling, P. F. Conforti, B. J. Garrison
Appl. Phys. A 79 (2004) 757
9. Coarse Grained Chemical Reaction Model
Y. G. Yingling and B. J. Garrison
J. Phys. Chem. B 108 (2004) 1815
-
-
- Feature Article
-
8. Photochemical Ablation of Organic Solids
Y. G. Yingling and B. J. Garrison
Nucl. Instrum. Methods Phys. Research B 203 (2003) 188
7. Computer Simulation of Laser Ablation of Molecular Substrates
L. V. Zhigilei, E. Leveugle, B. J. Garrison, Y. G. Yingling, M. I. Zeifman
Chemical Reviews 103 (2003) 321
6. MD Simulations of MALDesorption – Connections to Experiment
L. V. Zhigilei, Y. G. Yingling, T. E. Itina, T. A. Schoolcraft, B. J. Garrison
Int. J. Mass Spectrom. Ion Processes 226 (2003) 85
5. Big Molecule Ejection – SIMS vs. MALDI
B. J. Garrison, A. Delcorte, L. V. Zhigilei, T. E. Itina, K. D. Krantzman, Y. G. Yingling, C. M. McQuaw, E. J. Smiley, N. Winograd
Applied Surface Science 69-71 (2003) 203
4. Photochemical induced effects in material ejection in laser ablation
Y. G. Yingling and B. J. Garrison
Chem. Phys. Lett. 364 (2002) 237
3. The Role of the Photochemical Fragmentation in Laser Ablation
Y. G. Yingling, L. V. Zhigilei, B. J. Garrison
J. Photochemistry and Photobiology A: Chemistry 145 (2001) 173
2. Photochemical Fragmentation Processes in Laser Ablation of Organic Solids
Y. G. Yingling, L. V. Zhigilei, B. J. Garrison
Nucl. Instrun. Methods Phys. Research B 180 (2001) 171
1. Laser Ablation of bicomponent Systems: A Probe of Molecular Ejection Mechanisms
Y. G. Yingling, L. V. Zhigilei, B. J. Garrison, A. Koubenakis, J. Labrakis, S. Georgiou
Appl. Phys. Lett. 78 (2001) 1631