Products
Software (Machine learning, Analysis and Methods)
Codes are deposited into Yingling group public repository on GitHub https://github.com/yingling-group
- flory_fox_scraper Program to (a) scrape information on glass transition temperatures of polymers and (b) perform a Flory-Fox fit.
- Reference: J. S. Peerless, N. J. B. Milliken, M. D. Manning, T. J. Oweida, Y. G. Yingling, Advanced Theory and Simulations (2018) DOI: 10.1002/adts.201800129
- lammps-mspin LAMMPS plugin for magnetic nanoparticles simulation with atomistic resolution
- Reference: Akhlak U. Mahmood and Yaroslava G. Yingling, ACS Journal of Chemical Theory and Computations (2022) DOI: 10.1021/acs.jctc.1c01253
- X3DBio2: A Visual Analysis Tool for Biomolecular Structure Comparison
H. Yi, S. Thakur, L. Sethaphong, Y. G. Yingling
Proc. SPIE 8654, VDA (2013) 86540W doi:10.1117/12.2002626 - X3DBio: A Visual Analysis Tool for Biomolecular Structure Exploration
H. Yi, A. Singh, Y. G. Yingling
Proc. SPIE 8294 (2012) 82940S-1-8
Methods
All-Atom Simulation Method for Zeeman Alignment and Dipolar Assembly of Magnetic Nanoparticles
Akhlak U. Mahmood and Yaroslava G. Yingling
ACS Journal of Chemical Theory and Computations (2022) DOI: 10.1021/acs.jctc.1c01253
An Implicit Solvent Ionic Strength (ISIS) Method to Model Polyelectrolyte Systems with Dissipative Particle Dynamics
N. K. Li, W. H. Fuss, Y. G. Yingling
Macromolecular Theory and Simulations 24 (2015) 7-12.
Coarse-Grained Model of the Interaction of Light with Polymeric Material: Onset of Ablation
Y. G. Yingling, B. J. Garrison
Journal of Physical Chemistry B 109 (2005) 16482-16489
Coarse Grained Chemical Reaction Model
Y. G. Yingling and B. J. Garrison
J. Phys. Chem. B 108 (2004) 1815
Predicted 3D structures
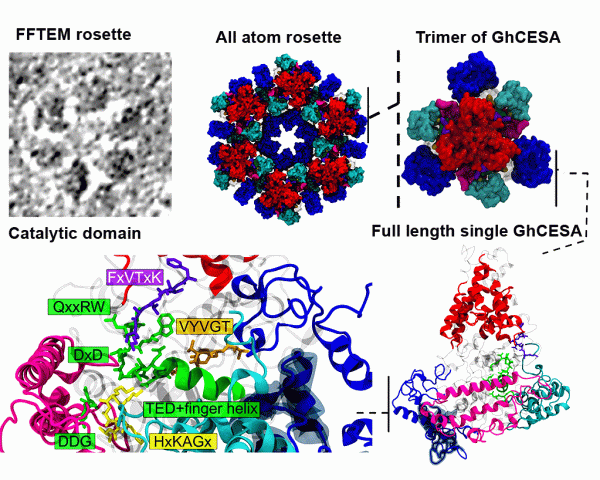 |
In silico structure prediction of full-length cotton cellulose synthase protein (GhCESA1) and its hierarchical complexes, Abhishek Singh, Albert L. Kwansa, Ho Shin Kim, Justin T. Williams, Hui Yang, Nan K. Li, James D. Kubicki, Alison W. Roberts, Candace H. Haigler, Yaroslava G. Yingling Cellulose 27 (2020) 5597-5616 DOI: 10.1007/s10570-020-03194-7 |
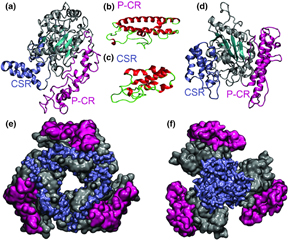 |
Prediction of the structures of the plant-specific regions of vascular plant cellulose synthases and correlated functional analysis, Latsavongsakda Sethaphong, Jonathan K. Davis, Erin Slabaugh, Abhishek Singh, Candace H. Haigler, Yaroslava G. Yingling Cellulose 23 (2016) 145-161 DOI: 10.1007/s10570-015-0789-6 |
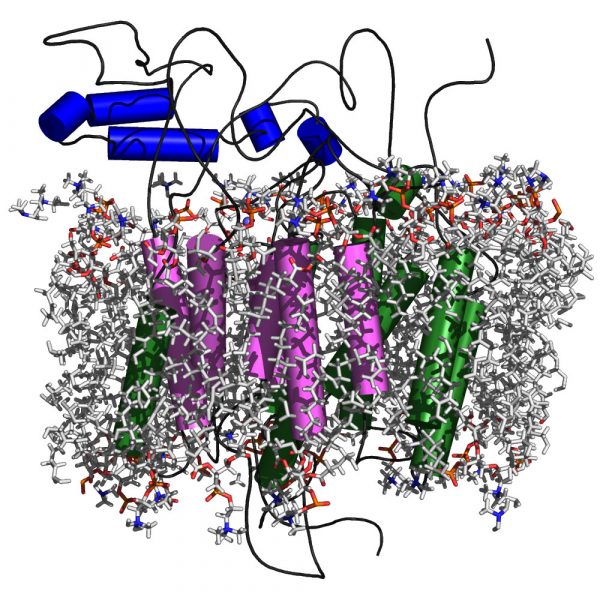 |
Na+ Dependent Anion Transport by a Barley Efflux Protein Revealed through an Integrative Platform, Yagnesh Nagarajan, Jay Rongala, Sukanya Luang, Abhishek Singh, Nadim Shadiac, Julie Hayes, Tim Sutton, Matthew Gilliham, Stephen Tyerman, Gordon McPhee, Nicolas H. Voelcker, Hayden D. T. Mertens, Nigel Kirby, Jung-Goo Lee, Yaroslava G. Yingling, Maria Hrmova Plant Cell 28 (2016) 202-218 DOI: 10.1105/tpc.15.00625 |
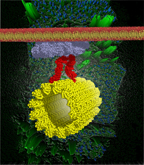 |
Cellulose synthase interactive 1 is required for a fast recycling of cellulose synthase complexes to the plasma membrane in Arabidopsis, Lei Lei, Abhishek Singh, Logan Bashline, Shundai Li, Yaroslava G. Yingling, and Ying Gu Plant Cell (2015) DOI: 10.1105/tpc.15.00442 |
- Tertiary model of a plant cellulose synthase
L. Sethaphong, C. H. Haigler, J. D. Kubicki, J. Zimmer, D. Bonetta, S. DeBolt, Y. G. Yingling
Proc. Natl. Acad. Sci. USA 110 (2013) 7512-7517 - The 3′ Proximal Translational Enhancer of Turnip Crinkle Virus Binds to 60S Ribosomal Subunits
V. A. Stupina, A. Meskauskas, J. C. Mccormack, Y. G. Yingling, B. A. Shapiro, J. D. Dinman, A. E. Simon
RNA 14 (2008) 2379-2393 - Structural Domains Within the 3′ UTR of Turnip Crinkle Virus
J. C. McCormack, X. Yuan, Y. G. Yingling, W. Kasprzak, R. E. Zamora, B. A. Shapiro, A. E. Simon
Journal of Virology 82 (2008) 8706-8720 - Computational design of an RNA hexagonal nano-ring and an RNA nanotube
Y. G. Yingling, B. A. Shapiro
Nano Letters 7 (2007) 2328-2334
Database
RNAJunction: a database of RNA junctions and kissing loops for three-dimensional structural analysis and nanodesign
E.Bindewald, R. Hayes, Y. G. Yingling, W. Kasprzak, B. A. Shapiro
Nucleic Acids Res. 36 (2008) D392-D397
- Link to RNAJunction database
Patents
- K. Clendennen, Y. G. Yingling, H. Kim, (2018), CALB Variants, US010035995 B2 WO2017100240 A1. The invention relates to amino acid sequence variants of a lipase with improved activity for catalyzing synthesis reactions and methods of preparing the variants. The methods include predicting amino acid sites for change based on computational models of the protein structure in non-aqueous conditions, and expressing the protein in a prokaryotic host for subsequent purification and use. The enzyme sequence variants described have a three to nine-fold improvement in synthesis activity over the parent protein sequence.
- A. Shapiro, Y. G. Yingling, E. Bindewald, W. Kasprzak, L. Jaeger, I. Severcan, C. Geary, K. Afonin (2010), RNA Nanoparticles and methods of use, WO2010148085-A1, US 20120263648 A1. The presently-disclosed subject matter relates to an artificial RNA nanostructure and method of use thereof. In particular, the presently-disclosed subject matter relates to RNA nanoparticles and RNA dendrimers, and methods of disease diagnosis and treatments using the RNA nanostructure and RNA dendrimers.
- G. Yingling, B. A. Shapiro (2007), RNA nanoparticles and nanotubes. EP2035043-A2; AU2007300734-A1; CA2654174-A1; US2010016409-A1. The instant invention provides polyvalent RNA nanoparticles comprising RNA motifs as building blocks that can form RNA nanotubes. The polyvalent RNA nanoparticles are suitable for therapeutic or diagnostic use in a number of diseases or disorders.